###############################################################################
# The Institute for the Design of Advanced Energy Systems Integrated Platform
# Framework (IDAES IP) was produced under the DOE Institute for the
# Design of Advanced Energy Systems (IDAES).
#
# Copyright (c) 2018-2023 by the software owners: The Regents of the
# University of California, through Lawrence Berkeley National Laboratory,
# National Technology & Engineering Solutions of Sandia, LLC, Carnegie Mellon
# University, West Virginia University Research Corporation, et al.
# All rights reserved. Please see the files COPYRIGHT.md and LICENSE.md
# for full copyright and license information.
###############################################################################
Supercritical CO2 Property Surrogate with PySMO Surrogate Object - flowsheet_optimization (Part 3)#
Maintainer: Javal Vyas
Author: Javal Vyas
Updated: 2024-01-24
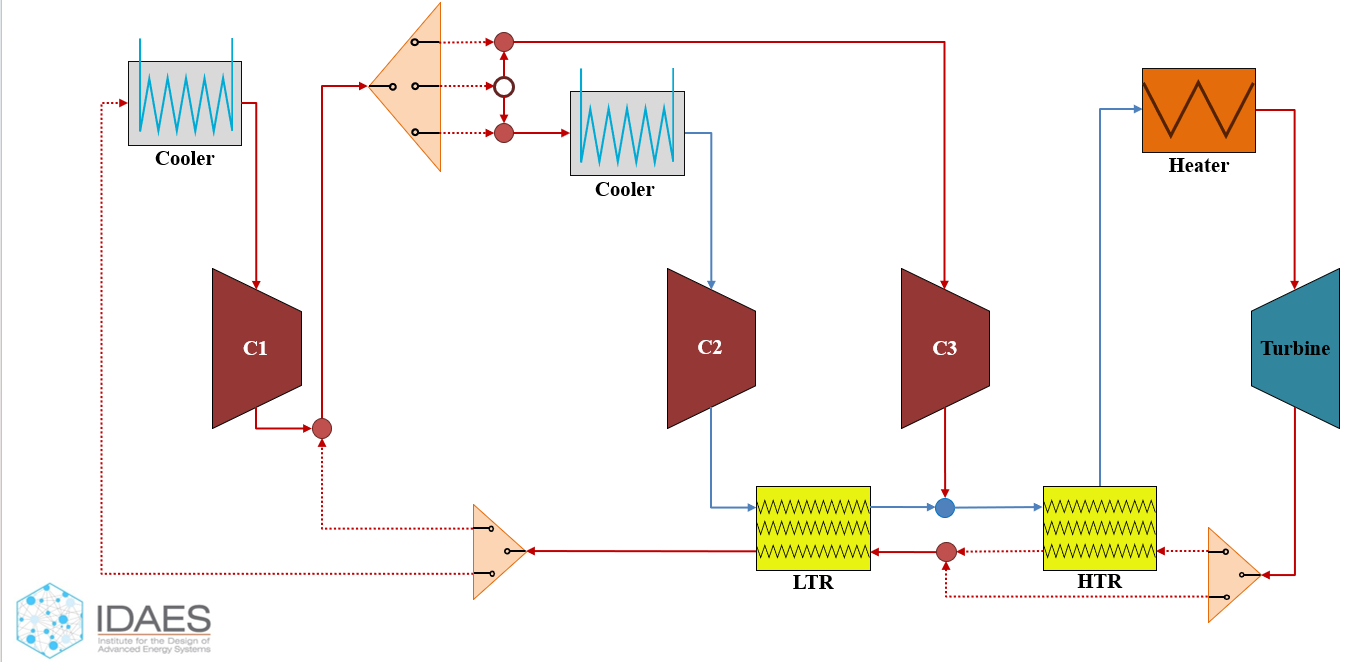
With the surrogate model being embedded in the property package, it is ready to be used in the flowsheet. We start by creating the following flowsheet using the IDAES package.
from IPython.display import Image
from pathlib import Path
def datafile_path(name):
return Path("..") / name
Image(datafile_path("CO2_flowsheet.png"))
1. Importing libraries#
We will be using the unit models from the IDAES package along with components from pyomo.environ and pyomo.network.
from pyomo.environ import (
ConcreteModel,
Block,
Var,
Param,
Constraint,
SolverFactory,
TransformationFactory,
TerminationCondition,
value,
Expression,
minimize,
units,
)
from pyomo.network import Arc, SequentialDecomposition
# Import IDAES libraries
from idaes.core import FlowsheetBlock, UnitModelBlockData
from idaes.models.unit_models import (
Mixer,
MomentumMixingType,
PressureChanger,
Heater,
Separator,
HeatExchanger,
)
from idaes.models.unit_models.pressure_changer import ThermodynamicAssumption
from idaes.core.util.model_statistics import degrees_of_freedom
from idaes.core.util.initialization import propagate_state
from properties import SCO2ParameterBlock
import idaes.logger as idaeslog
_log = idaeslog.getModelLogger("my_model", level=idaeslog.DEBUG, tag="model")
2. Constructing the flowsheet#
To construct the flowsheet we need to define a ConcreteModel using pyomo and then add a FlowsheetBlock to the ConcreteModel. Here since we are focusing on the steady state process, we shall have the dynamic flag as False in the FlowsheetBlock. Next, we define the properties in the FlowsheetBlock that we imported from the properties.py file. Then start adding the unit models to the FlowsheetBlock with the suitable arguments, after which we connect them using Arcs as in the flowsheet above.
Once we have the connected flowsheet, we initialize individual unit models. Before initializing, we fix desired variables for the desired behavior of the unit model and then use propagate_state to pass on the state variables to next unit model in the flowsheet. After completely initializing the flowsheet, we convert the network to a mathematical form by using network.expand_arcs from the TransformationFactory and apply it on the flowsheet block. Then we call the solver and solve the flowsheet to calculate the total work in the process.
def main():
# Setup solver and options
solver = SolverFactory("ipopt")
outlvl = 0
tee = True
# Set up concrete model
m = ConcreteModel()
# Create a flowsheet block
m.fs = FlowsheetBlock(dynamic=False)
# Create the properties param block
m.fs.properties = SCO2ParameterBlock()
# Add unit models to the flowsheet
m.fs.boiler = Heater(
dynamic=False, property_package=m.fs.properties, has_pressure_change=True
)
m.fs.turbine = PressureChanger(
dynamic=False,
property_package=m.fs.properties,
compressor=False,
thermodynamic_assumption=ThermodynamicAssumption.isentropic,
)
m.fs.HTR_pseudo_shell = Heater(
dynamic=False, property_package=m.fs.properties, has_pressure_change=True
)
m.fs.HTR_pseudo_tube = Heater(
dynamic=False, property_package=m.fs.properties, has_pressure_change=True
)
m.fs.LTR_pseudo_shell = Heater(
dynamic=False, property_package=m.fs.properties, has_pressure_change=True
)
m.fs.LTR_pseudo_tube = Heater(
dynamic=False, property_package=m.fs.properties, has_pressure_change=True
)
m.fs.splitter_1 = Separator(
property_package=m.fs.properties, outlet_list=["bypass", "to_cooler"]
)
m.fs.co2_cooler = Heater(
dynamic=False, property_package=m.fs.properties, has_pressure_change=True
)
m.fs.main_compressor = PressureChanger(
dynamic=False,
property_package=m.fs.properties,
compressor=True,
thermodynamic_assumption=ThermodynamicAssumption.isentropic,
)
m.fs.bypass_compressor = PressureChanger(
dynamic=False,
property_package=m.fs.properties,
compressor=True,
thermodynamic_assumption=ThermodynamicAssumption.isentropic,
)
m.fs.splitter_2 = Separator(
property_package=m.fs.properties,
ideal_separation=False,
outlet_list=["to_FG_cooler", "to_LTR"],
)
m.fs.FG_cooler = Heater(
dynamic=False, property_package=m.fs.properties, has_pressure_change=True
)
m.fs.mixer = Mixer(
property_package=m.fs.properties, inlet_list=["FG_out", "LTR_out", "bypass"]
)
# # Connect the flowsheet
m.fs.s01 = Arc(source=m.fs.boiler.outlet, destination=m.fs.turbine.inlet)
m.fs.s02 = Arc(source=m.fs.turbine.outlet, destination=m.fs.HTR_pseudo_shell.inlet)
m.fs.s03 = Arc(
source=m.fs.HTR_pseudo_shell.outlet, destination=m.fs.LTR_pseudo_shell.inlet
)
m.fs.s04 = Arc(
source=m.fs.LTR_pseudo_shell.outlet, destination=m.fs.splitter_1.inlet
)
m.fs.s05 = Arc(source=m.fs.splitter_1.to_cooler, destination=m.fs.co2_cooler.inlet)
m.fs.s06 = Arc(
source=m.fs.splitter_1.bypass, destination=m.fs.bypass_compressor.inlet
)
m.fs.s07 = Arc(
source=m.fs.co2_cooler.outlet, destination=m.fs.main_compressor.inlet
)
m.fs.s08 = Arc(source=m.fs.bypass_compressor.outlet, destination=m.fs.mixer.bypass)
m.fs.s09 = Arc(
source=m.fs.main_compressor.outlet, destination=m.fs.splitter_2.inlet
)
m.fs.s10 = Arc(
source=m.fs.splitter_2.to_FG_cooler, destination=m.fs.FG_cooler.inlet
)
m.fs.s11 = Arc(
source=m.fs.splitter_2.to_LTR, destination=m.fs.LTR_pseudo_tube.inlet
)
m.fs.s12 = Arc(source=m.fs.LTR_pseudo_tube.outlet, destination=m.fs.mixer.LTR_out)
m.fs.s13 = Arc(source=m.fs.FG_cooler.outlet, destination=m.fs.mixer.FG_out)
m.fs.s14 = Arc(source=m.fs.mixer.outlet, destination=m.fs.HTR_pseudo_tube.inlet)
# NETL Baseline
m.fs.boiler.inlet.flow_mol.fix(121.1)
m.fs.boiler.inlet.temperature.fix(685.15)
m.fs.boiler.inlet.pressure.fix(34.51)
m.fs.boiler.outlet.temperature.fix(893.15) # Turbine inlet T = 620 C
m.fs.boiler.deltaP.fix(-0.21)
m.fs.boiler.initialize(outlvl=outlvl)
propagate_state(m.fs.s01)
m.fs.turbine.ratioP.fix(1 / 3.68)
m.fs.turbine.efficiency_isentropic.fix(0.927)
m.fs.turbine.initialize(outlvl=outlvl)
propagate_state(m.fs.s02)
m.fs.HTR_pseudo_shell.outlet.temperature.fix(489.15)
m.fs.HTR_pseudo_shell.deltaP.fix(-0.07)
m.fs.HTR_pseudo_shell.initialize(outlvl=outlvl)
propagate_state(m.fs.s03)
m.fs.LTR_pseudo_shell.outlet.temperature.fix(354.15)
m.fs.LTR_pseudo_shell.deltaP.fix(-0.07)
m.fs.LTR_pseudo_shell.initialize(outlvl=outlvl)
propagate_state(m.fs.s04)
m.fs.splitter_1.split_fraction[0, "bypass"].fix(0.25)
m.fs.splitter_1.initialize(outlvl=outlvl)
propagate_state(m.fs.s05)
m.fs.co2_cooler.outlet.temperature.fix(308.15)
m.fs.co2_cooler.deltaP.fix(-0.07)
m.fs.co2_cooler.initialize(outlvl=outlvl)
propagate_state(m.fs.s06)
m.fs.bypass_compressor.efficiency_isentropic.fix(0.85)
m.fs.bypass_compressor.ratioP.fix(3.8)
m.fs.bypass_compressor.initialize(outlvl=outlvl)
propagate_state(m.fs.s07)
m.fs.main_compressor.efficiency_isentropic.fix(0.85)
m.fs.main_compressor.ratioP.fix(3.8)
m.fs.main_compressor.initialize(outlvl=outlvl)
propagate_state(m.fs.s09)
m.fs.splitter_2.split_fraction[0, "to_FG_cooler"].fix(0.046)
m.fs.splitter_2.initialize(outlvl=outlvl)
propagate_state(m.fs.s10)
m.fs.FG_cooler.outlet.temperature.fix(483.15)
m.fs.FG_cooler.deltaP.fix(-0.06)
m.fs.FG_cooler.initialize(outlvl=outlvl)
propagate_state(m.fs.s11)
m.fs.LTR_pseudo_tube.deltaP.fix(0)
m.fs.LTR_pseudo_tube.heat_duty[0].fix(-value(m.fs.LTR_pseudo_shell.heat_duty[0]))
m.fs.LTR_pseudo_tube.initialize(outlvl=outlvl)
# Add constraint heats of the LTR_pseudo shell and tube
m.fs.LTR_pseudo_tube.heat_duty[0].unfix()
m.fs.c1 = Constraint(
expr=m.fs.LTR_pseudo_shell.heat_duty[0] == -m.fs.LTR_pseudo_tube.heat_duty[0]
)
propagate_state(m.fs.s08)
propagate_state(m.fs.s12)
propagate_state(m.fs.s13)
m.fs.mixer.initialize(outlvl=outlvl)
propagate_state(m.fs.s14)
m.fs.HTR_pseudo_tube.heat_duty[0].fix(-value(m.fs.HTR_pseudo_shell.heat_duty[0]))
m.fs.HTR_pseudo_tube.deltaP.fix(-0.07)
m.fs.HTR_pseudo_tube.initialize(outlvl=outlvl)
m.fs.HTR_pseudo_tube.heat_duty[0].unfix()
m.fs.c2 = Constraint(
expr=m.fs.HTR_pseudo_shell.heat_duty[0] == -m.fs.HTR_pseudo_tube.heat_duty[0]
)
TransformationFactory("network.expand_arcs").apply_to(m.fs)
print("--------------------------------------------------------------------")
print("The degrees of freedom for the flowsheet is ", degrees_of_freedom(m))
print("--------------------------------------------------------------------")
solver.solve(m, tee=tee)
#
from idaes.core.util.units_of_measurement import (
convert_quantity_to_reporting_units,
report_quantity,
)
# Print reports
for i in m.fs.component_objects(Block):
if isinstance(i, UnitModelBlockData):
i.report()
# Converting units for readability
print(
-1 * value(units.convert(m.fs.turbine.work_mechanical[0], units.kW))
- 1 * value(units.convert(m.fs.main_compressor.work_mechanical[0], units.kW))
- 1 * value(units.convert(m.fs.bypass_compressor.work_mechanical[0], units.kW)),
units.kW,
)
return m
if __name__ == "__main__":
m = main()
2023-08-19 23:45:27 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234527.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234527.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:27 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234527.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234527.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:27 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234527.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234527.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:27 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234527.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234527.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.core.surrogate.pysmo_surrogate: Decode surrogate. type=poly
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
===========================Polynomial Regression===============================================
Warning: solution.pickle already exists; results will be saved to " solution_v_08-19-23_234528.pickle ".
The number of cross-validation cases (3) is used.
The default training/cross-validation split of 0.75 is used.
No iterations will be run.
Default parameter estimation method is used.
Parameter estimation method: pyomo
2023-08-19 23:45:28 [INFO] idaes.init.fs.boiler.control_volume: Initialization Complete
2023-08-19 23:45:28 [INFO] idaes.init.fs.boiler: Initialization Complete: optimal - Optimal Solution Found
2023-08-19 23:45:29 [INFO] idaes.init.fs.turbine: Initialization Complete: optimal - Optimal Solution Found
2023-08-19 23:45:29 [INFO] idaes.init.fs.HTR_pseudo_shell.control_volume: Initialization Complete
2023-08-19 23:45:29 [INFO] idaes.init.fs.HTR_pseudo_shell: Initialization Complete: optimal - Optimal Solution Found
2023-08-19 23:45:29 [INFO] idaes.init.fs.LTR_pseudo_shell.control_volume: Initialization Complete
2023-08-19 23:45:29 [INFO] idaes.init.fs.LTR_pseudo_shell: Initialization Complete: optimal - Optimal Solution Found
2023-08-19 23:45:29 [INFO] idaes.init.fs.splitter_1: Initialization Step 2 Complete: optimal - Optimal Solution Found
2023-08-19 23:45:29 [INFO] idaes.init.fs.co2_cooler.control_volume: Initialization Complete
2023-08-19 23:45:29 [INFO] idaes.init.fs.co2_cooler: Initialization Complete: optimal - Optimal Solution Found
2023-08-19 23:45:30 [INFO] idaes.init.fs.bypass_compressor: Initialization Complete: optimal - Optimal Solution Found
2023-08-19 23:45:30 [INFO] idaes.init.fs.main_compressor: Initialization Complete: optimal - Optimal Solution Found
2023-08-19 23:45:30 [INFO] idaes.init.fs.splitter_2: Initialization Step 2 Complete: optimal - Optimal Solution Found
2023-08-19 23:45:30 [INFO] idaes.init.fs.FG_cooler.control_volume: Initialization Complete
2023-08-19 23:45:30 [INFO] idaes.init.fs.FG_cooler: Initialization Complete: optimal - Optimal Solution Found
2023-08-19 23:45:30 [INFO] idaes.init.fs.LTR_pseudo_tube.control_volume: Initialization Complete
2023-08-19 23:45:30 [INFO] idaes.init.fs.LTR_pseudo_tube: Initialization Complete: optimal - Optimal Solution Found
2023-08-19 23:45:30 [INFO] idaes.init.fs.mixer: Initialization Complete: optimal - Optimal Solution Found
2023-08-19 23:45:30 [INFO] idaes.init.fs.HTR_pseudo_tube.control_volume: Initialization Complete
2023-08-19 23:45:31 [INFO] idaes.init.fs.HTR_pseudo_tube: Initialization Complete: optimal - Optimal Solution Found
--------------------------------------------------------------------
The degrees of freedom for the flowsheet is 0
--------------------------------------------------------------------
Ipopt 3.13.2:
******************************************************************************
This program contains Ipopt, a library for large-scale nonlinear optimization.
Ipopt is released as open source code under the Eclipse Public License (EPL).
For more information visit http://projects.coin-or.org/Ipopt
This version of Ipopt was compiled from source code available at
https://github.com/IDAES/Ipopt as part of the Institute for the Design of
Advanced Energy Systems Process Systems Engineering Framework (IDAES PSE
Framework) Copyright (c) 2018-2019. See https://github.com/IDAES/idaes-pse.
This version of Ipopt was compiled using HSL, a collection of Fortran codes
for large-scale scientific computation. All technical papers, sales and
publicity material resulting from use of the HSL codes within IPOPT must
contain the following acknowledgement:
HSL, a collection of Fortran codes for large-scale scientific
computation. See http://www.hsl.rl.ac.uk.
******************************************************************************
This is Ipopt version 3.13.2, running with linear solver ma27.
Number of nonzeros in equality constraint Jacobian...: 452
Number of nonzeros in inequality constraint Jacobian.: 0
Number of nonzeros in Lagrangian Hessian.............: 118
Total number of variables............................: 178
variables with only lower bounds: 32
variables with lower and upper bounds: 59
variables with only upper bounds: 0
Total number of equality constraints.................: 178
Total number of inequality constraints...............: 0
inequality constraints with only lower bounds: 0
inequality constraints with lower and upper bounds: 0
inequality constraints with only upper bounds: 0
iter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls
0 0.0000000e+00 1.12e+02 1.00e+00 -1.0 0.00e+00 - 0.00e+00 0.00e+00 0
1 0.0000000e+00 3.28e-01 1.12e-02 -1.0 1.32e+01 - 9.89e-01 1.00e+00h 1
2 0.0000000e+00 5.45e-06 1.05e-06 -1.0 1.32e+01 - 1.00e+00 1.00e+00h 1
3 0.0000000e+00 1.37e-08 2.83e-08 -2.5 2.87e-04 - 1.00e+00 1.00e+00h 1
Number of Iterations....: 3
(scaled) (unscaled)
Objective...............: 0.0000000000000000e+00 0.0000000000000000e+00
Dual infeasibility......: 0.0000000000000000e+00 0.0000000000000000e+00
Constraint violation....: 3.4924596548080444e-10 1.3737007975578308e-08
Complementarity.........: 0.0000000000000000e+00 0.0000000000000000e+00
Overall NLP error.......: 3.4924596548080444e-10 1.3737007975578308e-08
Number of objective function evaluations = 4
Number of objective gradient evaluations = 4
Number of equality constraint evaluations = 4
Number of inequality constraint evaluations = 0
Number of equality constraint Jacobian evaluations = 4
Number of inequality constraint Jacobian evaluations = 0
Number of Lagrangian Hessian evaluations = 3
Total CPU secs in IPOPT (w/o function evaluations) = 0.004
Total CPU secs in NLP function evaluations = 0.002
EXIT: Optimal Solution Found.
====================================================================================
Unit : fs.boiler Time: 0.0
------------------------------------------------------------------------------------
Unit Performance
Variables:
Key : Value : Units : Fixed : Bounds
Heat Duty : 1.4382e+06 : watt : False : (None, None)
------------------------------------------------------------------------------------
Stream Table
Units Inlet Outlet
flow_mol mole / second 1.2110e+05 1.2110e+05
temperature kelvin 685.15 893.15
pressure pascal 3.4510e+07 3.4300e+07
====================================================================================
====================================================================================
Unit : fs.turbine Time: 0.0
------------------------------------------------------------------------------------
Unit Performance
Variables:
Key : Value : Units : Fixed : Bounds
Isentropic Efficiency : 0.92700 : dimensionless : True : (None, None)
Mechanical Work : -9.9927e+05 : watt : False : (None, None)
Pressure Change : -24.979 : pascal : False : (None, None)
Pressure Ratio : 0.27174 : dimensionless : True : (None, None)
------------------------------------------------------------------------------------
Stream Table
Units Inlet Outlet
flow_mol mole / second 1.2110e+05 1.2110e+05
temperature kelvin 893.15 729.38
pressure pascal 3.4300e+07 9.3207e+06
====================================================================================
====================================================================================
Unit : fs.HTR_pseudo_shell Time: 0.0
------------------------------------------------------------------------------------
Unit Performance
Variables:
Key : Value : Units : Fixed : Bounds
Heat Duty : -1.4056e+06 : watt : False : (None, None)
------------------------------------------------------------------------------------
Stream Table
Units Inlet Outlet
flow_mol mole / second 1.2110e+05 1.2110e+05
temperature kelvin 729.38 489.15
pressure pascal 9.3207e+06 9.2507e+06
====================================================================================
====================================================================================
Unit : fs.HTR_pseudo_tube Time: 0.0
------------------------------------------------------------------------------------
Unit Performance
Variables:
Key : Value : Units : Fixed : Bounds
Heat Duty : 1.4056e+06 : watt : False : (None, None)
------------------------------------------------------------------------------------
Stream Table
Units Inlet Outlet
flow_mol mole / second 1.2110e+05 1.2110e+05
temperature kelvin 535.47 736.02
pressure pascal 3.4560e+07 3.4490e+07
====================================================================================
====================================================================================
Unit : fs.LTR_pseudo_shell Time: 0.0
------------------------------------------------------------------------------------
Unit Performance
Variables:
Key : Value : Units : Fixed : Bounds
Heat Duty : -1.0929e+06 : watt : False : (None, None)
------------------------------------------------------------------------------------
Stream Table
Units Inlet Outlet
flow_mol mole / second 1.2110e+05 1.2110e+05
temperature kelvin 489.15 354.15
pressure pascal 9.2507e+06 9.1807e+06
====================================================================================
====================================================================================
Unit : fs.LTR_pseudo_tube Time: 0.0
------------------------------------------------------------------------------------
Unit Performance
Variables:
Key : Value : Units : Fixed : Bounds
Heat Duty : 1.0929e+06 : watt : False : (None, None)
------------------------------------------------------------------------------------
Stream Table
Units Inlet Outlet
flow_mol mole / second 86647. 86647.
temperature kelvin 378.99 566.32
pressure pascal 3.4620e+07 3.4620e+07
====================================================================================
====================================================================================
Unit : fs.splitter_1 Time: 0.0
------------------------------------------------------------------------------------
Unit Performance
Variables:
Key : Value : Units : Fixed : Bounds
Split Fraction [('bypass',)] : 0.25000 : dimensionless : True : (None, None)
Split Fraction [('to_cooler',)] : 0.75000 : dimensionless : False : (None, None)
------------------------------------------------------------------------------------
Stream Table
Units Inlet bypass to_cooler
flow_mol mole / second 1.2110e+05 30275. 90825.
temperature kelvin 354.15 354.15 354.15
pressure pascal 9.1807e+06 9.1807e+06 9.1807e+06
====================================================================================
====================================================================================
Unit : fs.co2_cooler Time: 0.0
------------------------------------------------------------------------------------
Unit Performance
Variables:
Key : Value : Units : Fixed : Bounds
Heat Duty : -4.4513e+05 : watt : False : (None, None)
------------------------------------------------------------------------------------
Stream Table
Units Inlet Outlet
flow_mol mole / second 90825. 90825.
temperature kelvin 354.15 308.15
pressure pascal 9.1807e+06 9.1107e+06
====================================================================================
====================================================================================
Unit : fs.main_compressor Time: 0.0
------------------------------------------------------------------------------------
Unit Performance
Variables:
Key : Value : Units : Fixed : Bounds
Isentropic Efficiency : 0.85000 : dimensionless : True : (None, None)
Mechanical Work : 2.2092e+05 : watt : False : (None, None)
Pressure Change : 25.510 : pascal : False : (None, None)
Pressure Ratio : 3.8000 : dimensionless : True : (None, None)
------------------------------------------------------------------------------------
Stream Table
Units Inlet Outlet
flow_mol mole / second 90825. 90825.
temperature kelvin 308.15 378.99
pressure pascal 9.1107e+06 3.4620e+07
====================================================================================
====================================================================================
Unit : fs.bypass_compressor Time: 0.0
------------------------------------------------------------------------------------
Unit Performance
Variables:
Key : Value : Units : Fixed : Bounds
Isentropic Efficiency : 0.85000 : dimensionless : True : (None, None)
Mechanical Work : 1.1041e+05 : watt : False : (None, None)
Pressure Change : 25.706 : pascal : False : (None, None)
Pressure Ratio : 3.8000 : dimensionless : True : (None, None)
------------------------------------------------------------------------------------
Stream Table
Units Inlet Outlet
flow_mol mole / second 30275. 30275.
temperature kelvin 354.15 460.04
pressure pascal 9.1807e+06 3.4886e+07
====================================================================================
====================================================================================
Unit : fs.splitter_2 Time: 0.0
------------------------------------------------------------------------------------
Unit Performance
Variables:
Key : Value : Units : Fixed : Bounds
Split Fraction [('to_FG_cooler',)] : 0.046000 : dimensionless : True : (None, None)
Split Fraction [('to_LTR',)] : 0.95400 : dimensionless : False : (None, None)
------------------------------------------------------------------------------------
Stream Table
Units Inlet to_FG_cooler to_LTR
flow_mol mole / second 90825. 4177.9 86647.
temperature kelvin 378.99 378.99 378.99
pressure pascal 3.4620e+07 3.4620e+07 3.4620e+07
====================================================================================
====================================================================================
Unit : fs.FG_cooler Time: 0.0
------------------------------------------------------------------------------------
Unit Performance
Variables:
Key : Value : Units : Fixed : Bounds
Heat Duty : 31903. : watt : False : (None, None)
------------------------------------------------------------------------------------
Stream Table
Units Inlet Outlet
flow_mol mole / second 4177.9 4177.9
temperature kelvin 378.99 483.15
pressure pascal 3.4620e+07 3.4560e+07
====================================================================================
====================================================================================
Unit : fs.mixer Time: 0.0
------------------------------------------------------------------------------------
Stream Table
Units FG_out LTR_out bypass Outlet
flow_mol mole / second 4177.9 86647. 30275. 1.2110e+05
temperature kelvin 483.15 566.32 460.04 535.47
pressure pascal 3.4560e+07 3.4620e+07 3.4886e+07 3.4560e+07
====================================================================================
667.9424945058901 kW

